
The algorithm simplifies the analysis of multiple sequence data by condensing the mass of information present, and thus allows the rapid identification of substitutions of structural and functional importance. Group 3 download - AV Manager Digital Display Software 9. The DNA sequence of interest is used as a template for a special type of PCR called chain-termination PCR. The analysis of charge conservation across annexin domains identifies the locations at which conserved charges change sign. There are three main steps to Sanger sequencing. The algorithm is illustrated by application to an alignment of 67 SH2 domains where patterns of conserved hydrophobic residues that constitute the protein core are highlighted. ADDED Sequence Names can be modified directly in the Sequence Window using a contextual menu available by CTRL-Clicking (or Right-clicking) on the Sequence.
The algorithm is encoded in the computer program AMAS (Analysis of Multiply Aligned Sequences) which provides a textual summary of the analysis and an annotated (boxed, shaded and/or coloured) multiple sequence alignment. All pairs of subgroups are then compared to highlight positions that confer the unique features of each subgroup.
Serial cloner two sequences serial#
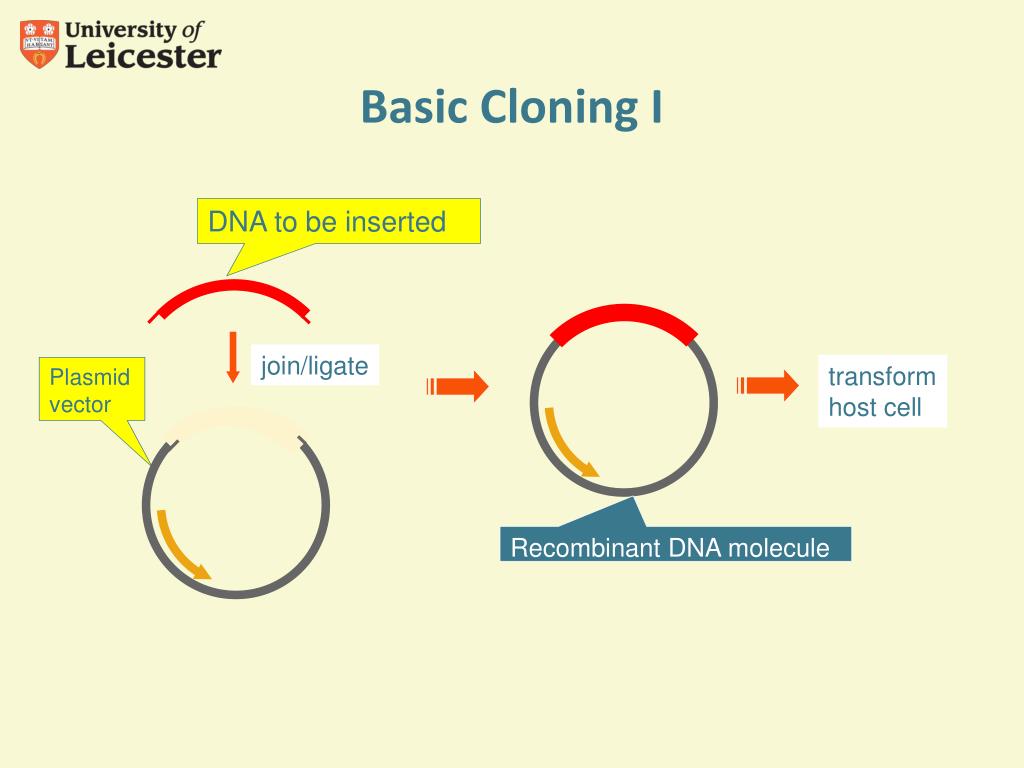
Sequences in the alignment are gathered into subgroups on the basis of sequence similarity, functional, evolutionary or other criteria. Sequence usage in MM-BIR and template grants 1F32GM096690, R01GM076020, and 5R37GM020056, and switching were carried out by either the alignment feature work in the Petes laboratory is supported by NIH/NIHMS grants contained in the Serial Cloner 2.6.1 (Franck Perez, Serialbasics) R01GM24410 and RO1GM52319. You can show or hide the Help window using a menu command available in the Window menu and on the Toolbar window. A general description is also available inside Serial Cloner using the built-in Help window. The strategy is based on a flexible set-based description of amino acid properties, which is used to define the conservation between any group of amino acids. functions of Serial Cloner as well as some useful tips. Figure 2.In the first step the DNA of interest (red) in the donor plasmid needs to be cut out. The new algorithm allows questions important in the design of mutagenesis experiments to be quickly answered since positions in the alignment that show unusual or interesting residue substitution patterns may be rapidly identified. This enables you to work with sequences created by other laboratories in order to compare the results. An algorithm is described for the systematic characterization of the physico-chemical properties seen at each position in a multiple protein sequence alignment. Background Identifying nuclease-induced double-stranded breaks in DNA on a genome-wide scale is critical for assessing the safety and efficacy of genome editing therapies. MacVector, ApE, DNAstar, pDRAW32 and GenBank.


 0 kommentar(er)
0 kommentar(er)
